Particle dynamics
Recurrent Neural Nets as a Particle Dynamics Integrator
The second IU initial application shows a rather different type of surrogate and illustrates an SBI goal to collect benchmarks covering a range of surrogate designs. Molecular dynamics simulations rely on numerical integrators such as Verlet to solve Newton’s equations of motion. Using a sufficiently small time step to avoid discretization errors, Verlet integrators generate a trajectory of particle positions as solutions to the equations of motions. In 1, 2, 3, the IU team introduces an integrator based on recurrent neural networks that is trained on trajectories generated using the Verlet integrator and learns to propagate the dynamics of particles with timestep up to 4000 times larger compared to the Verlet timestep. As shown in Fig. 4 (right) the error does not increase as one evolves the system for the surrogate while standard Verlet integration in Fig. 4 (left) has unacceptable errors even for time steps of just 10 times that used in an accurate simulation. The surrogate demonstrates a significant net speedup over Verlet of up to 32000 for few-particle (1 - 16) 3D systems and over a variety of force fields including the Lennard-Jones (LJ) potential. This application uses a recurrent plus dense neural network architecture and illustrates an important approach to learning evolution operators which can be applied across a variety of fields including Earthquake science (IU work in progress) and Fusion 4.
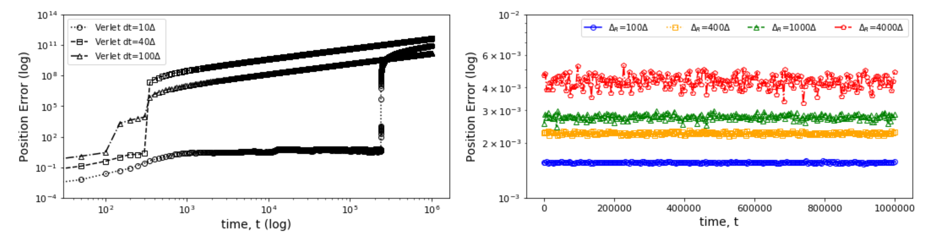
Fig. 4: Average error in position updates for 16 particles interacting with an LJ potential, The left figure is standard MD with error increasing for ∆t as 10, 40, or 100 times robust choice (0.001). On the right is the LSTM network with modest error up to t = 106 even for ∆t = 4000 times the robust MD choice.
References
JCS Kadupitiya, Geoffrey C. Fox, Vikram Jadhao, “GitHub repository for Simulating Molecular Dynamics with Large Timesteps using Recurrent Neural Networks.” [Online]. Available: https://github.com/softmaterialslab/RNN-MD. [Accessed: 01-May-2020] ↩︎
J. C. S. Kadupitiya, G. C. Fox, and V. Jadhao, “Simulating Molecular Dynamics with Large Timesteps using Recurrent Neural Networks,” arXiv [physics.comp-ph], 12-Apr-2020 [Online]. Available: http://arxiv.org/abs/2004.06493 ↩︎
J. C. S. Kadupitiya, G. Fox, and V. Jadhao, “Recurrent Neural Networks Based Integrators for Molecular Dynamics Simulations,” in APS March Meeting 2020, 2020 [Online]. Available: http://meetings.aps.org/Meeting/MAR20/Session/L45.2. [Accessed: 23-Feb-2020] ↩︎
J. Kates-Harbeck, A. Svyatkovskiy, and W. Tang, “Predicting disruptive instabilities in controlled fusion plasmas through deep learning,” Nature, vol. 568, no. 7753, pp. 526–531, Apr. 2019 [Online]. Available: https://doi.org/10.1038/s41586-019-1116-4 ↩︎