A list of surrogates
This is the multi-page printable view of this section. Click here to print.
Surrogates
- 1: AutoPhaseNN: unsupervised physics-aware deep learning of 3D nanoscale Bragg coherent diffraction imaging
- 2: Calorimeter surrogates
- 3: Virtual tissue
- 4: Cosmoflow
- 5: Fully ionized plasma fluid model closures
- 6: Ions in nanoconfinement
- 7: Molecule docking
- 8: miniWeatherML
- 9: OSMI
- 10: Particle dynamics
- 11: Performance Surrogate
- 12: PtychoNN: deep learning network for ptychographic imaging that predicts sample amplitude and phase from diffraction data.
1 - AutoPhaseNN: unsupervised physics-aware deep learning of 3D nanoscale Bragg coherent diffraction imaging
Metadata
Model autophasenn.json
Datasets autoPhaseNN_aicdi.json
AutoPhaseNN 1, a physics-aware unsupervised deep convolutional neural network (CNN) that learns to solve the phase problem without ever being shown real space images of the sample amplitude or phase. By incorporating the physics of the X-ray scattering into the network design and training, AutoPhaseNN learns to predict both the amplitude and phase of the sample given the measured diffraction intensity alone. Additionally, unlike previous deep learning models, AutoPhaseNN does not need the ground truth images of sample’s amplitude and phase at any point, either in training or in deployment. Once trained, the physical model is discarded and only the CNN portion is needed which has learned the data inversion from reciprocal space to real space and is ~100 times faster than the iterative phase retrieval with comparable image quality. Furthermore, we show that by using AutoPhaseNN’s prediction as the learned prior to iterative phase retrieval, we can achieve consistently higher image quality, than neural network prediction alone, at 10 times faster speed than iterative phase retrieval alone.
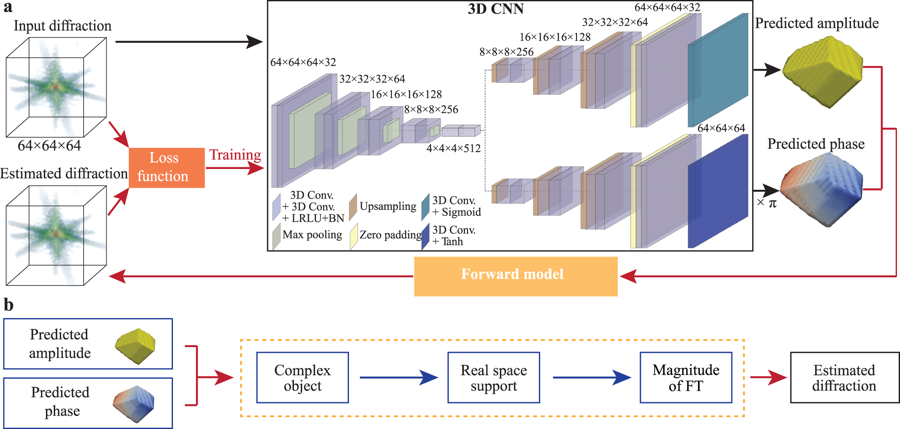
Fig. 1: Schematic of the neural network structure of AutoPhaseNN model during training.
a) The model consists of a 3D CNN and the X-ray scattering forward model. The 3D
CNN is implemented with a convolutional auto-encoder and two deconvolutional
decoders using the convolutional, maximum pooling, upsampling and zero padding
layers. The physical knowledge is enforced via the Sigmoid and Tanh activation
functions in the final layers. b The X-ray scattering forward model includes the
numerical modeling of diffraction and the image shape constraints. It takes the
amplitude and phase from the 3D CNN output to form the complex image. Then the
estimated diffraction pattern is obtained from the FT of the current estimation
of the real space image.
Image from: Yao, Y. et al / CC-BY
References
Yao, Y., Chan, H., Sankaranarayanan, S. et al. AutoPhaseNN: unsupervised physics-aware deep learning of 3D nanoscale Bragg coherent diffraction imaging. npj Comput Mater 8, 124 (2022). https://doi.org/10.1038/s41524-022-00803-w ↩︎ ↩︎
2 - Calorimeter surrogates
Overview
The Kaggle calorimeter challenge 1 2 uses generative AI to produce a surrogate for the Monte Carlo calculation of a calorimeter response to an incident particle (ATLAS data at LHC calculated with GEANT4). Variational Auto Encoders, GANs, Normalizing Flows, and Diffusion Models. We also have a surrogate using a Quantum Computer (DWAVE) annealer to generate random samples. We have identified four different surrogates that are available openly from Kaggle and later submissions.
References
3 - Virtual tissue
Overview
Neural networks (NNs) have been demonstrated to be a viable alternative to traditional direct numerical evaluation algorithms, with the potential to accelerate computational time by several orders of magnitude. In the present paper we study the use of encoder-decoder convolutional neural network (CNN) algorithms as surrogates for steady-state diffusion solvers. The construction of such surrogates requires the selection of an appropriate task, network architecture, training set structure and size, loss function, and training algorithm hyperparameters. It is well known that each of these factors can have a significant impact on the performance of the resultant model. Our approach employs an encoder-decoder CNN architecture, which we posit is particularly wellsuited for this task due to its ability to effectively transform data, as opposed to merely compressing it. We systematically evaluate a range of loss functions, hyperparameters, and training set sizes. Our results indicate that increasing the size of the training set has a substantial effect on reducing performance fluctuations and overall error. Additionally, we observe that the performance of the model exhibits a logarithmic dependence on the training set size. Furthermore, we investigate the effect on model performance by using different subsets of data with varying features. Our results highlight the importance of sampling the configurational space in an optimal manner, as this can have a significant impact on the performance of the model and the required training time. In conclusion, our results suggest that training a model with a pre-determined error performance bound is not a viable approach, as it does not guarantee that edge cases with errors larger than the bound do not exist. Furthermore, as most surrogate tasks involve a high dimensional landscape, an ever increasing training set size is, in principle, needed, however it is not a practical solution.
References
https://arxiv.org/pdf/2302.03786.pdf is virtual tissue ↩︎
There is an earlier surrogate referred to in this arxiv. It was published https://www.frontiersin.org/articles/10.3389/fphys.2021.667828/full ↩︎
4 - Cosmoflow
Metadata
Model cosmoflow.json
Datasets
cosmoUniverse_2019_05_4parE_tf_v2.json
cosmoUniverse_2019_05_4parE_tf_v2_mini.json
Overview
This application is based on the original CosmoFlow paper presented at SC18 and continued by the ExaLearn project, and adopted as a benchmark in the MLPerf HPC suite. It involves training a 3D convolutional neural network on N-body cosmology simulation data to predict physical parameters of the universe. The reference implementation for MLPerf HPC v0.5 CosmoFlow uses TensorFlow with the Keras API and Horovod for data-parallel distributed training. The dataset comes from simulations run by ExaLearn, with universe volumes split into cubes of size 128x128x128 with 4 redshift bins. The total dataset volume preprocessed for MLPerf HPC v0.5 in TFRecord format is 5.1 TB. The target objective in MLPerf HPC v0.5 is to train the model to a validation mean-average-error < 0.124. However, the problem size can be scaled down and the training throughput can be used as the primary objective for a small scale or shorter timescale benchmark.123
References
5 - Fully ionized plasma fluid model closures
Fully ionized plasma fluid model closures (Argonne):1 The closure problem in fluid modeling is a well-known challenge to modelers aiming to accurately describe their system of interest. Analytic formulations in a wide range of regimes exist but a practical, generalized fluid closure for magnetized plasmas remains an elusive goal. There are scenarios where complex physics prevents a simple closure being assumed, and the question as to what closure to employ has a non-trivial answer. In a proof-of-concept study, Argonne researchers turned to machine learning to try to construct surrogate closure models that map the known macroscopic variables in a fluid model to the higher-order moments that must be closed. In their study, the researchers considered three closures: Braginskii, Hammett-Perkins, and Guo-Tang; for each of them, they tried three types of ANNs: locally connected, convolutional, and fully connected. Applying a physics-informed machine learning approach, they found that there is a benefit to tailoring a specific network architecture informed by the physics of the plasma regime each closure is designed for, rather than carelessly applying an unnecessarily complex general network architecture. will choose one of the surrogates and bring it up an early example for SBI with reference implementation and tutorial documentation. As a follow-up, the Argonne team will tackle more challenging problems.
References
R. Maulik, N. A. Garland, X.-Z. Tang, and P. Balaprakash, “Neural network representability of fully ionized plasma fluid model closures,” arXiv [physics.comp-ph], 10-Feb-2020 [Online]. Available: http://arxiv.org/abs/2002.04106 ↩︎
6 - Ions in nanoconfinement
Metadata
Model nanoconfinement.json
Datasets nanoconfinement.json
This application 1 2 3 studies ionic structure in electrolyte solutions in nanochannels with planar uncharged surfaces and can use multiple molecular dynamics (MD) codes including LAMMPS 4 which run on HPC supercomputers with OpenMP and MPI parallelization.
A dense neural-net (NN) was used to learn 150 final state characteristics based on the input of 5 parameters with typical results shown in fig. 2(b) with the NN results for three important densities tracking well the MD simulation results for a wide range of unseen input system parameters. Fig. 3(a,b) shows two typical density profiles with again the NN prediction tracking well the simulation. Input quantities were confinement length, positive ion valency, negative ion valency, salt concentration, and ion diameter. Figure 2(a) shows the runtime architecture for dynamic use and update of the NN and our middleware discussed in Sec. 3.2.6 will generalize this. The inference time for this on a single core is 104 times faster than the parallel code which is itself 100 times the sequential code. This surrogate approach is the first-of-its-kind in the area of simulating charged soft-matter systems and there are many other published papers in both biomolecular and material science presenting similar successful surrogates 5 with a NN architecture similar to fig. 3(c).
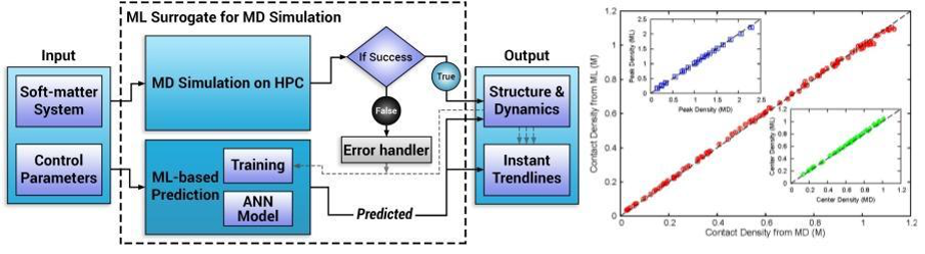
Fig. 2 a) Architecture of dynamic training of ML surrogate and b) Comparison of three final state densities (peak, contact, and center) between MD simulations and NN surrogate predictions [^5] [^51].
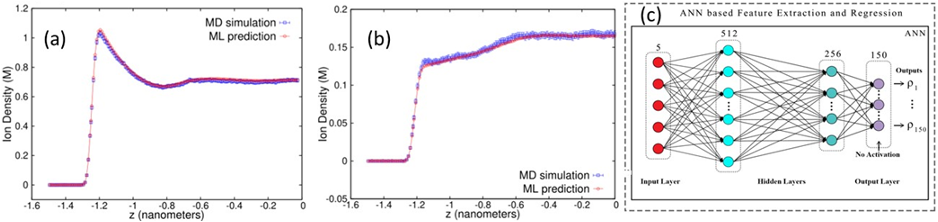
Fig. 3 (a,b) Two density profiles of confined ions for very different input parameters and comparing MD and NN. (c) Fully connected deep learning network used to learn the final densities. ReLU activation units are in the 512 and 256 node hidden layers. The output values were learned on 150 nodes.
References
JCS Kadupitiya , Geoffrey C. Fox , and Vikram Jadhao, “Machine learning for performance enhancement of molecular dynamics simulations,” in International Conference on Computational Science ICCS2019, Faro, Algarve, Portugal, 2019 [Online]. Available: http://dsc.soic.indiana.edu/publications/ICCS8.pdf ↩︎
J. C. S. Kadupitiya, F. Sun, G. Fox, and V. Jadhao, “Machine learning surrogates for molecular dynamics simulations of soft materials,” J. Comput. Sci., vol. 42, p. 101107, Apr. 2020 [Online]. Available: http://www.sciencedirect.com/science/article/pii/S1877750319310609 ↩︎
“Molecular Dynamics for Nanoconfinement.” [Online]. Available: https://github.com/softmaterialslab/nanoconfinement-md. [Accessed: 11-May-2020] ↩︎
S. Plimpton, “Fast Parallel Algorithms for Short Range Molecular Dynamics,” J. Comput. Phys., vol. 117, pp. 1–19, 1995 [Online]. Available: http://faculty.chas.uni.edu/~rothm/Modeling/Parallel/Plimpton.pdf ↩︎
Geoffrey Fox, Shantenu Jha, “Learning Everywhere: A Taxonomy for the Integration of Machine Learning and Simulations,” in IEEE eScience 2019 Conference, San Diego, California [Online]. Available: https://arxiv.org/abs/1909.13340 ↩︎
7 - Molecule docking
Molecule docking: Docking small molecules to a protein’s binding site is often one of the first steps for virtual screening 1. Although many open-source and commercial packages exist for docking, AI approaches can be equally powerful (and computationally more efficient) for docking studies 2. Utilizing advances in control from reinforcement learning (RL), Argonne researchers trained an agent to drive the docking of a rigid ligand into a flexible protein pocket. The RL agent treats the ligand as a rigid body to which it can move through affine transformations along the protein. This procedure bypasses sampling on a grid as the agent is trained to optimize the pose against OpenEye Fred docking function 3, and/or other openly available docking tools such as UCSF DOCK, Autodock/Vina. The challenge of this approach is that there is a need to train the agent based on the protein target, which can still take considerable time on single-GPU systems. This area comes from the major Argonne CANDLE 4 project and other applications (DeepDriveMD) will come from this project in the new submissions category.
Refernces
P. D. Lyne, “Structure-based virtual screening: an overview,” Drug Discov. Today, vol. 7, no. 20, pp. 1047–1055, Oct. 2002 [Online]. Available: http://dx.doi.org/10.1016/s1359-6446(02)02483-2 ↩︎
J. Li, A. Fu, and L. Zhang, “An overview of scoring functions used for protein–ligand interactions in molecular docking,” Interdiscip. Sci., pp. 1–9, 2019 [Online]. Available: https://idp.springer.com/authorize/casa?redirect_uri=https://link.springer.com/article/10.1007/s12539-019-00327-w&casa_token=Usuqtf4tu-4AAAAA:VD0uKAo49lSwaEEpmufft87cpUtbmE9MSdlR_Wpv880jHArsLIfLy8PQPAaN6ODJIArQ9GMz15wJ6lSX ↩︎
M. McGann, “FRED pose prediction and virtual screening accuracy,” J. Chem. Inf. Model., vol. 51, no. 3, pp. 578–596, Mar. 2011 [Online]. Available: http://dx.doi.org/10.1021/ci100436p ↩︎
“CANDLE Exascale Deep Learning and Simulation Enabled Precision Medicine for Cancer.” [Online]. Available: https://candle.cels.anl.gov/. [Accessed: 01-May-2020] ↩︎
8 - miniWeatherML
Metadata
Model miniWeatherML.json
Datasets miniWeatherML.json
Overview
MiniWeatherML is a playground for learning and developing Machine Learning (ML) surrogate models and workflows. It is based on a simplified weather model simulating flows such as supercells that are realistic enough to be challenging and simple enough for rapid prototyping in:
- Data generation and curation
- Machine Learning model training
- ML model deployment and analysis
- End-to-end workflows
References
9 - OSMI
Overview
We explore the relationship between certain network configurations and the performance of distributed Machine Learning systems. We build upon the Open Surrogate Model Inference (OSMI) Benchmark, a distributed inference benchmark for analyzing the performance of machine-learned surrogate models developed by Wes Brewer et. Al. We focus on analyzing distributed machine-learning systems, via machine-learned surrogate models, across varied hardware environments. By deploying the OSMI Benchmark on platforms like Rivanna HPC, WSL, and Ubuntu, we offer a comprehensive study of system performance under different configurations. The paper presents insights into optimizing distributed machine learning systems, enhancing their scalability and efficiency. We also develope a framework for automating the OSMI benchmark.
Introdcution
With the proliferation of machine learning as a tool for science, the need for efficient and scalable systems is paramount. This paper explores the Open Surrogate Model Inference (OSMI) Benchmark, a tool for testing the performance of machine-learning systems via machine-learned surrogate models. The OSMI Benchmark, originally created by Wes Brewer and colleagues, serves to evaluate various configurations and their impact on system performance.
Our research pivots around the deployment and analysis of the OSMI Benchmark across various hardware platforms, including the high-performance computing (HPC) system Rivanna, Windows Subsystem for Linux (WSL), and Ubuntu environments.
In each experiment, there are a variable number of TensorFlow model server instances, overseen by a HAProxy load balancer that distributes inference requests among the servers. Each server instance operates on a dedicated GPU, choosing between the V100 or A100 GPUs available on Rivanna. This setup mirrors real-world scenarios where load balancing is crucial for system efficiency.
On the client side, we initiate a variable number of concurrent clients executing the OSMI benchmark to simulate different levels of system load and analyze the corresponding inference throughput.
On top of the original OSMI-Bench, we implemented an object-oriented interface in Python for running experiments with ease, streamlining the process of benchmarking and analysis. The experiments rely on custom-built images based on NVIDIA’s tensorflow image. The code works on several hardwares, assuming the proper images are built.
Additionally, We develop a script for launching simultaneous experiments with permutations of pre-defined parameters with Cloudmesh Experiment-Executor. The Experiment Executor is a tool that automates the generation and execution of experiment variations with different parameters. This automation is crucial for conducting tests across a spectrum of scenarios.
Finally, we analyze the inference throughput and total time for each experiment. By graphing and examining these results, we draw critical insights into the performance dynamics of distributed machine learning systems.
In summary, a comprehensive examination of the OSMI Benchmark in diverse distributed ML systems is provided. We aim to contribute to the optimization of these systems, by providing a framework for finding the best performant system configuration for a given use case. Our findings pave the way for more efficient and scalable distributed computing environments.
The architectural view of the benchmarks are depictued in Figure 1 and Figure 2.
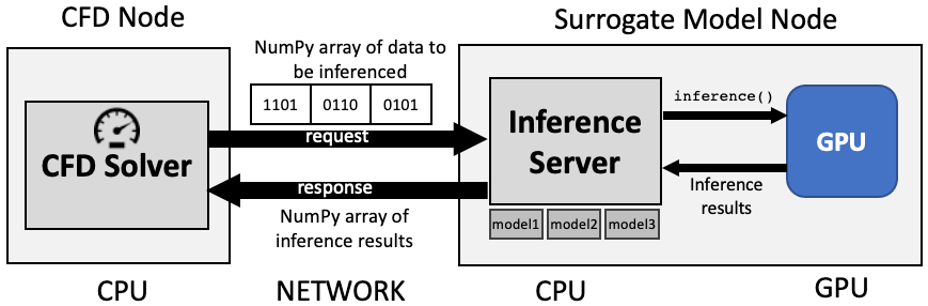
Figure 1: Surrogate calculations via a Inference Server.
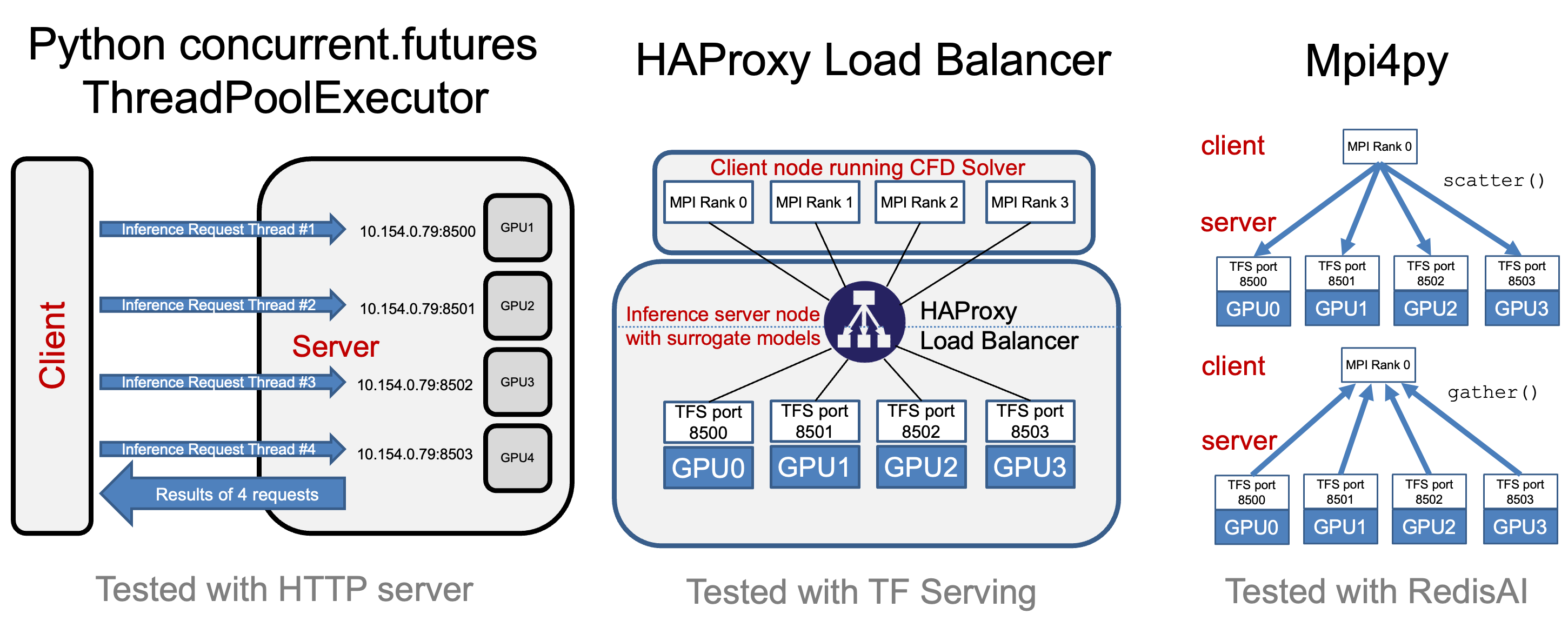
Figure 2: Possible benchmark configurations to measure sped of parallel iference.
References
Brewer, Wesley, Daniel Martinez, Mathew Boyer, Dylan Jude, Andy Wissink, Ben Parsons, Junqi Yin, and Valentine Anantharaj. “Production Deployment of Machine-Learned Rotorcraft Surrogate Models on HPC.” In 2021 IEEE/ACM Workshop on Machine Learning in High Performance Computing Environments (MLHPC), pp. 21-32. IEEE, 2021, https://ieeexplore.ieee.org/abstract/document/9652868, Note that OSMI-Bench differs from SMI-Bench described in the paper only in that the models that are used in OSMI are trained on synthetic data, whereas the models in SMI were trained using data from proprietary CFD simulations. Also, the OSMI medium and large models are very similar architectures as the SMI medium and large models, but not identical. ↩︎
Brewer, Wesley, Greg Behm, Alan Scheinine, Ben Parsons, Wesley Emeneker, and Robert P. Trevino. “iBench: a distributed inference simulation and benchmark suite.” In 2020 IEEE High Performance Extreme Computing Conference (HPEC), pp. 1-6. IEEE, 2020. ↩︎
Brewer, Wesley, Greg Behm, Alan Scheinine, Ben Parsons, Wesley Emeneker, and Robert P. Trevino. “Inference benchmarking on HPC systems.” In 2020 IEEE High Performance Extreme Computing Conference (HPEC), pp. 1-9. IEEE, 2020. ↩︎
Brewer, Wesley, Chris Geyer, Dardo Kleiner, and Connor Horne. “Streaming Detection and Classification Performance of a POWER9 Edge Supercomputer.” In 2021 IEEE High Performance Extreme Computing Conference (HPEC), pp. 1-7. IEEE, 2021. ↩︎
Gregor von Laszewski, J. P. Fleischer, and Geoffrey C. Fox. 2022. Hybrid Reusable Computational Analytics Workflow Management with Cloudmesh. https://doi.org/10.48550/ARXIV.2210.16941 ↩︎
10 - Particle dynamics
Recurrent Neural Nets as a Particle Dynamics Integrator
The second IU initial application shows a rather different type of surrogate and illustrates an SBI goal to collect benchmarks covering a range of surrogate designs. Molecular dynamics simulations rely on numerical integrators such as Verlet to solve Newton’s equations of motion. Using a sufficiently small time step to avoid discretization errors, Verlet integrators generate a trajectory of particle positions as solutions to the equations of motions. In 1 2 3, the IU team introduces an integrator based on recurrent neural networks that is trained on trajectories generated using the Verlet integrator and learns to propagate the dynamics of particles with timestep up to 4000 times larger compared to the Verlet timestep. As shown in Fig. 4 (right) the error does not increase as one evolves the system for the surrogate while standard Verlet integration in Fig. 4 (left) has unacceptable errors even for time steps of just 10 times that used in an accurate simulation. The surrogate demonstrates a significant net speedup over Verlet of up to 32000 for few-particle (1 - 16) 3D systems and over a variety of force fields including the Lennard-Jones (LJ) potential. This application uses a recurrent plus dense neural network architecture and illustrates an important approach to learning evolution operators which can be applied across a variety of fields including Earthquake science (IU work in progress) and Fusion 4.
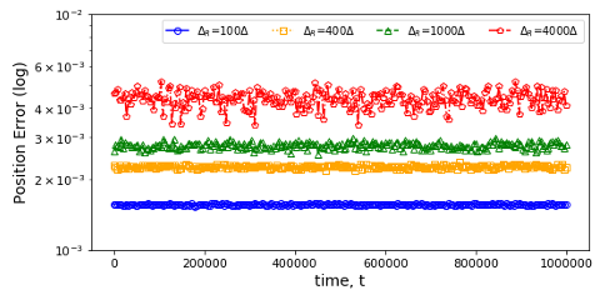
Fig. 4: Average error in position updates for 16 particles interacting with an LJ potential, The left figure is standard MD with error increasing for ∆t as 10, 40, or 100 times robust choice (0.001). On the right is the LSTM network with modest error up to t = 106 even for ∆t = 4000 times the robust MD choice.
Refernces
JCS Kadupitiya, Geoffrey C. Fox, Vikram Jadhao, “GitHub repository for Simulating Molecular Dynamics with Large Timesteps using Recurrent Neural Networks.” [Online]. Available: https://github.com/softmaterialslab/RNN-MD. [Accessed: 01-May-2020] ↩︎
J. C. S. Kadupitiya, G. C. Fox, and V. Jadhao, “Simulating Molecular Dynamics with Large Timesteps using Recurrent Neural Networks,” arXiv [physics.comp-ph], 12-Apr-2020 [Online]. Available: http://arxiv.org/abs/2004.06493 ↩︎
J. C. S. Kadupitiya, G. Fox, and V. Jadhao, “Recurrent Neural Networks Based Integrators for Molecular Dynamics Simulations,” in APS March Meeting 2020, 2020 [Online]. Available: http://meetings.aps.org/Meeting/MAR20/Session/L45.2. [Accessed: 23-Feb-2020] ↩︎
J. Kates-Harbeck, A. Svyatkovskiy, and W. Tang, “Predicting disruptive instabilities in controlled fusion plasmas through deep learning,” Nature, vol. 568, no. 7753, pp. 526–531, Apr. 2019 [Online]. Available: https://doi.org/10.1038/s41586-019-1116-4 ↩︎
11 - Performance Surrogate
Overview
Please contact Piotre
References
12 - PtychoNN: deep learning network for ptychographic imaging that predicts sample amplitude and phase from diffraction data.
Metadata
Model ptychonn.json
Datasets ptychonn_20191008_39.json
PtychoNN, uses a deep convolutional neural network to predict realspace structure and phase from far-field diffraction data. It recovers high fidelity amplitude and phase contrast images of a real sample hundreds of times faster than current ptychography reconstruction packages and reduces sampling requirements 1
References
Mathew J. Cherukara, Tao Zhou, Youssef Nashed, Pablo Enfedaque, Alex Hexemer, Ross J. Harder, Martin V. Holt; AI-enabled high-resolution scanning coherent diffraction imaging. Appl. Phys. Lett. 27 July 2020; 117 (4): 044103. https://doi.org/10.1063/5.0013065 ↩︎